A 3D printable microplate for distortion detection in expansion microscopy
Read more on our paper in Cell Reports Physical Science
Our team is pleased to share one of our latest projects in using a 3D printable microplate systems for (a) reducing the manual handling of hydrogels, (b) improving throughput, and (c) enabling convenient distortion checking of expansion microscopy (ExM) samples, particularly 4x (pro-) ExM. The idea of moving to a plate/array-based system is emerging fast; it comes from the slightly uncomfortable observation of the level of skill and care required to handle gels during an ExM protocol. These skills are absolutely trainable, a fact that is reinforced time and time again by hundreds of world class research groups that now use ExM to study the structure of complex samples. So many areas of the Life Sciences leverage plain skill and dexterity of researchers in preparing the primary samples and materials for the research. From the point of view of improving accessibility and reproducibility however, particular if you watch any of the practical demos of ExM like this video demo by the Synthetic Neurobiology Group, it can be seen that careful, yet direct, handling of the gels particularly between the gellation and imaging steps is necessary. If the structures that you aim to visualise are at the very bottom surface of the cells, handling and transferring the gel during the ExM protocol needs to be done with extreme care.
The concept is straightforward. To avoid picking up, transferring and cutting the gel, one needs a microscopy-ready chamber that can allow the casting of the gel within a mould. Removing the mould then allows the sample to expand into the rest of the well. If one makes the well a predictable shape with limited rotational symmetries, the orientation of the gel can be preserved from the beginning of the experiment to the post-ExM structure. Use a microplate that combine eight of these chamber units, and you get a multi-sample array that can fit on any standard microscope stage.
The plate based ExM approach may not be the perfect solution, however we have published our approach primarily with the view that alternative ways to standardise and improve the through-put of this technique if we would like to see broader uptake of the method.
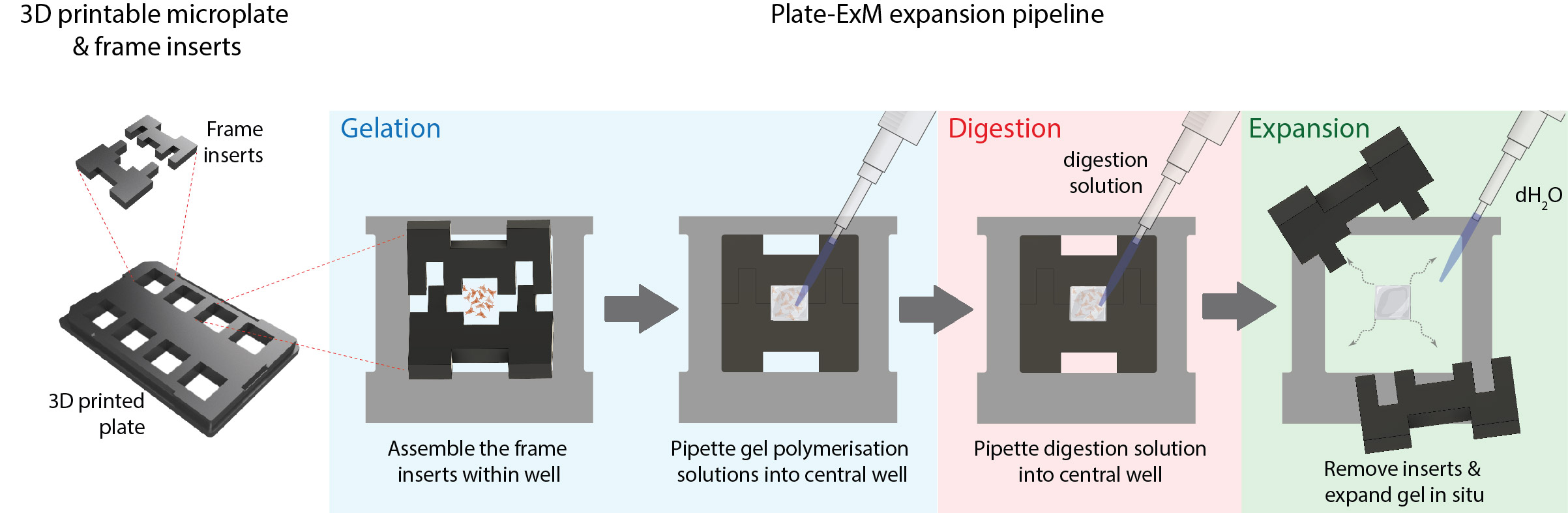
The concept is straightforward. To avoid picking up, transferring and cutting the gel, one needs a microscopy-ready chamber that can allow the casting of the gel within a mould. Removing the mould then allows the sample to expand into the rest of the well. If one makes the well a predictable shape with limited rotational symmetries, the orientation of the gel can be preserved from the beginning of the experiment to the post-ExM structure. Use a microplate that combine eight of these chamber units, and you get a multi-sample array that can fit on any standard microscope stage.
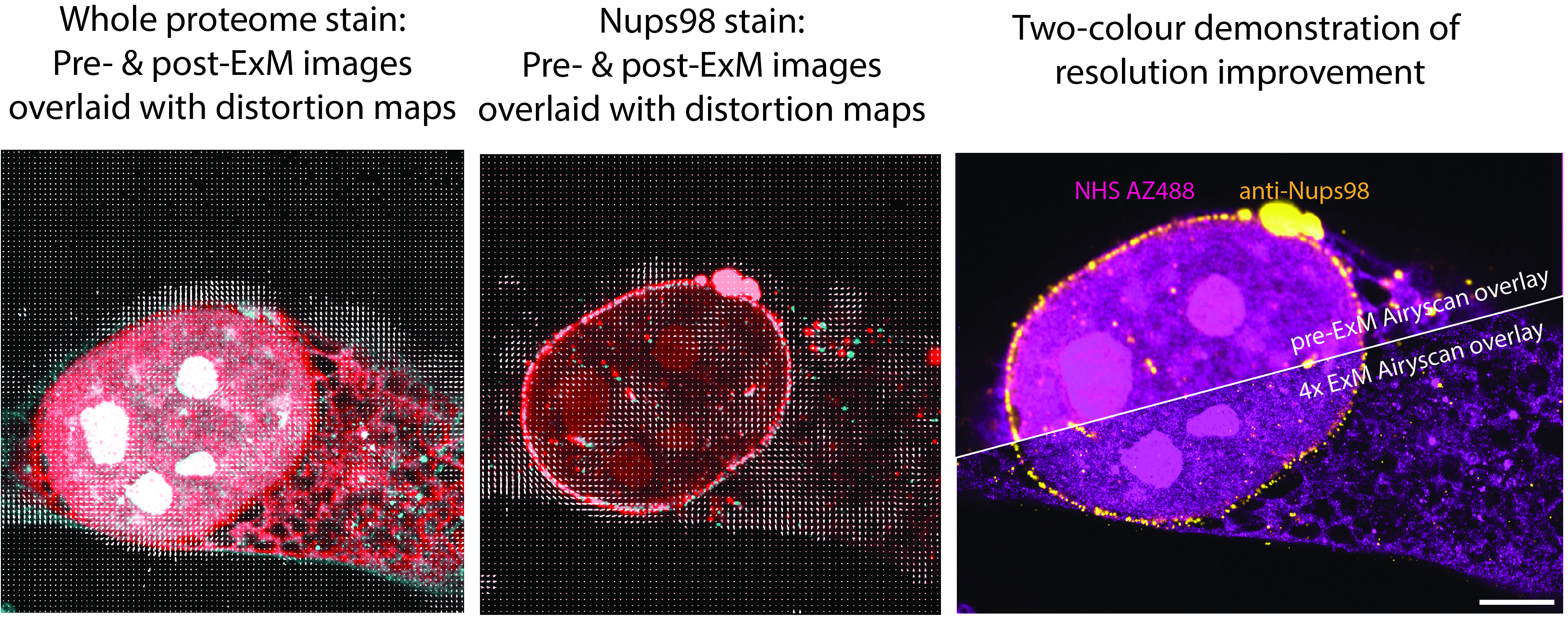
Some of the most interesting observations we made along the way were that most of the distortions were introduced during the digestion/denaturation step of the ExM protocol – something observed with three-point repeated imaging with the plate ExM. Whilst there was potential for using the B-spline registration vectors to ‘correct’ the distortions, we found that this often sacrificed the resolution gain achieved by ExM. So, at least in the short term, the “see and avoid” approach is a better approach when it comes to dealing with distortions in expansion microscopy.
You can view exemplar datasets and download the code and ImageJ macros we used for this from the data supplement at Zenodo.
