Projects & Resources
We have a number of projects on the go, and this page outlines some of the main areas of research at present. We are also passionate about developing and broadly sharing tools and technologies that we develop for our research. If these research areas and tech are of interest to you, either to come and work in our team or for collaboration, please get in touch.
Featured projects

In a 3-year project funded by New South Wales Medical Research, we are developing single molecule imaging and spatial omics tools to study systems of key signalling molecules and their regulators in the healthy and failing myocardium. We are bringing together multiplexed super-resolution imaging, big data pipelines and machine learning to understand how these molecular systems of signalling lose their balance as a heart begins to fail.

In a 7-year project funded by UK Research & Innovation, we are developing several innovations that are aimed at making super-resolution microscopy, expansion microscopy in particular, accessible and affordable for life sciences researchers. Amongst some of the work completed so far, we have characterised a series of 14 nondescript stains and 3D printed microplates for expansion microscopy. As a part of this project, we are also applying super-resolution to sample types and sub-fields that have not utilised super-resolution as much as mainstream cell biology. They include imaging thick tissues and whole organisms such as sea urchin larvae. More coming soon.
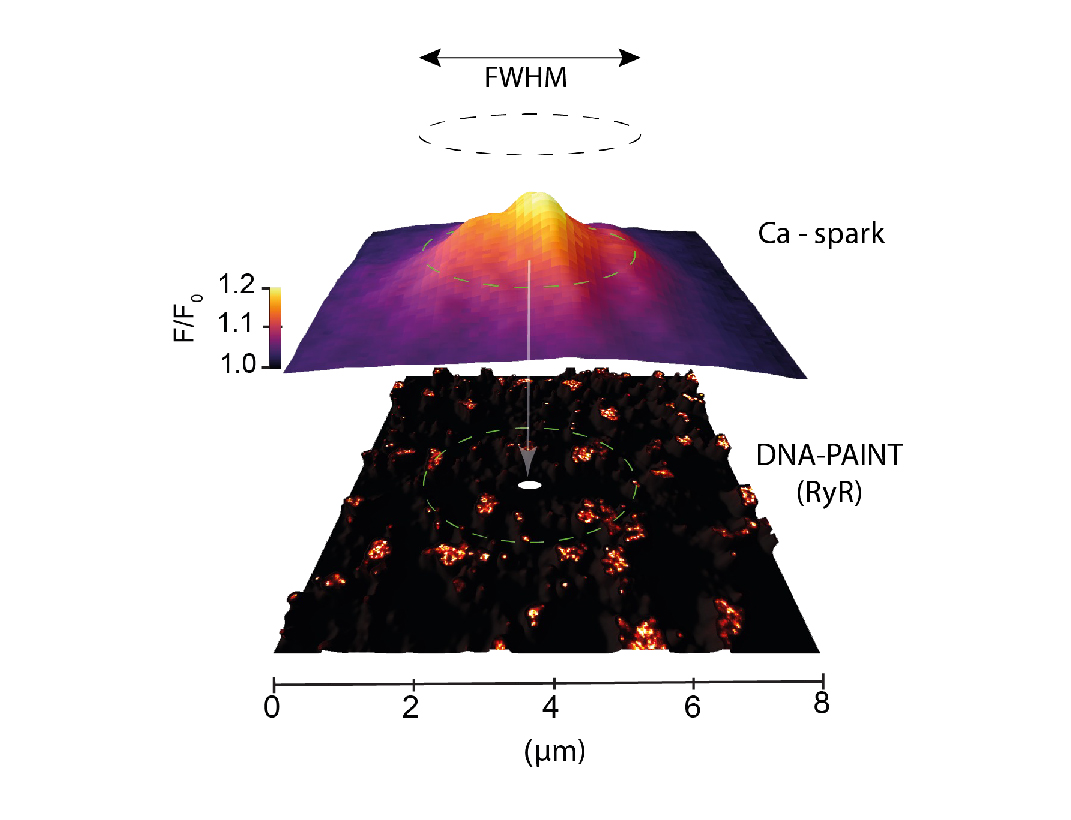
We have an ongoing interest in the calcium signals (calcium sparks) produced by intracellular ion channels such as ryanodine receptors and inositol triphosphate receptors. In cardiac muscle cells, ryanodine receptors organised into clusters within distinct signalling nanodomains work together in specific spatial patterns to produce these sparks. In pain sensing neurons from the dorsal root ganglia, these channels are less well clustered and produce much more variable sparks. Using a correlative imaging and analysis pipeline, we have pioneered the methods to visualising and decoding the spatial relationships between these calcium sparks and super-resolution maps of these channels.
Resources & tools
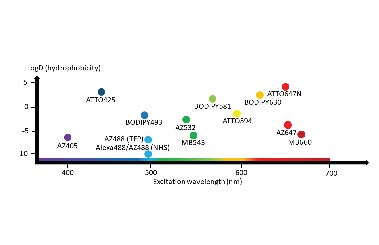
An ongoing project looks to characterise the diversity of staining patterns that afford weighted staining to specific or groups of cell-compartments looks to offer researchers a range of new options and guidance to leverage fluorescent ester dyes as counter-stains in diffraction-limited and expansion microscopy. See rently publication in the journal Nanoscale.
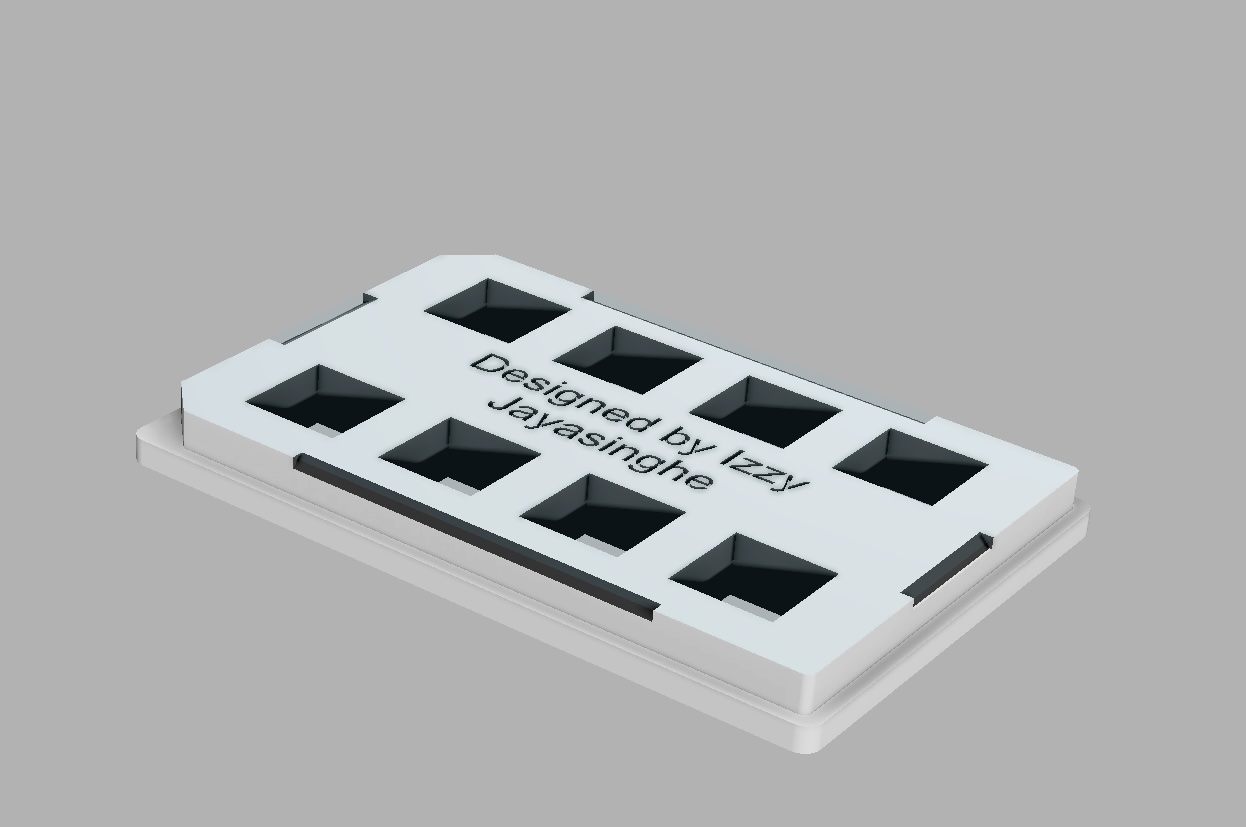
Read more about our latest projects in using a 3D printable microplate systems for (a) reducing the manual handling of hydrogels, (b) improving throughput, and (c) enabling convenient distortion checking of expansion microscopy (ExM) samples.